Receptor Mediated Endocytosis
Relating to work conducted from 2008 to 2011
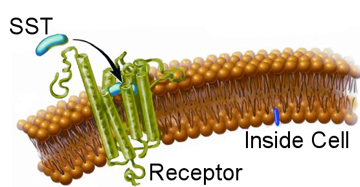 With a basic knowledge of how nano-particles enter cells explained in the nano-particles endocytosis section, the next step is to understand how proteins and cells interact. As discussed before, cells use a process known as receptor mediated endocytosis to gather material from the outside world. This can either be food for the cell or signalling proteins to regulate processes within the cell itself. Our final goal is to use nano-particles to track such signalling proteins. In the nano-particles endocytosis section we learned how to stop nano-particles entering cells non-specifically, here we wish to learn how to make them interact specifically. Again, due to the complexities of biology, we start with a simple system and gradually build up a full picture. We are interested in the peptide Somatostatin (SST for short), which does various jobs in the body including regulate blood pressure. When an SST molecule comes into contact with the corresponding receptor on the cells surface it will bind to it, as depicted in the image. This binding event causes a string of complex knock-on effects within the cell. Around the receptor itself other proteins will start to gather and stick to the cell membrane; in combination these will start to build a scaffold like structure. The structure is naturally curved and so as the scaffold grows it will try to form a sphere, the scaffold proteins are stronger than the cell membrane so this process is allowed to continue until the original receptor is sitting within a small bubble on the inside of the cell with the SST molecule that triggered these events on the inside of the bubble, which is known as a vesicle. Other proteins will also have been alerted to the presence of the SST molecule and the vesicle will be dragged along filaments within the cell to where it will be processed by yet other proteins. Finally, the SST molecule will be dismantled and the receptor will be sent back to the membrane of the process to begin again. While this is complex, the act of triggering the receptor also set off many other sets of protein reactions. For example, in the case of SST, the receptor also signalled to other proteins to open tiny gates in the cell membrane that only allow calcium ions through.
With a basic knowledge of how nano-particles enter cells explained in the nano-particles endocytosis section, the next step is to understand how proteins and cells interact. As discussed before, cells use a process known as receptor mediated endocytosis to gather material from the outside world. This can either be food for the cell or signalling proteins to regulate processes within the cell itself. Our final goal is to use nano-particles to track such signalling proteins. In the nano-particles endocytosis section we learned how to stop nano-particles entering cells non-specifically, here we wish to learn how to make them interact specifically. Again, due to the complexities of biology, we start with a simple system and gradually build up a full picture. We are interested in the peptide Somatostatin (SST for short), which does various jobs in the body including regulate blood pressure. When an SST molecule comes into contact with the corresponding receptor on the cells surface it will bind to it, as depicted in the image. This binding event causes a string of complex knock-on effects within the cell. Around the receptor itself other proteins will start to gather and stick to the cell membrane; in combination these will start to build a scaffold like structure. The structure is naturally curved and so as the scaffold grows it will try to form a sphere, the scaffold proteins are stronger than the cell membrane so this process is allowed to continue until the original receptor is sitting within a small bubble on the inside of the cell with the SST molecule that triggered these events on the inside of the bubble, which is known as a vesicle. Other proteins will also have been alerted to the presence of the SST molecule and the vesicle will be dragged along filaments within the cell to where it will be processed by yet other proteins. Finally, the SST molecule will be dismantled and the receptor will be sent back to the membrane of the process to begin again. While this is complex, the act of triggering the receptor also set off many other sets of protein reactions. For example, in the case of SST, the receptor also signalled to other proteins to open tiny gates in the cell membrane that only allow calcium ions through.
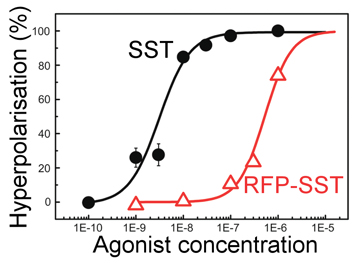 This in turn leads to a small change in the charge on the cell membrane, and this is a useful tool to check if SST is entering the cell. The plot, right, shows just such an experiment. Here SST is referred to as an agonist as it is the molecule responsible for setting off the cellular responses. The bottom axis shows the number of SST molecules added to a batch of cells and the y-axis shows the recorded level of cell hyperpolarisation - the amount of charge on the membrane (this is recoded using another dye where the amount of fluorescence relates to the local charge). The curve shown (black) is typical of what is to be expected if things are working well. At low concentrations there are not enough triggered SST receptors to create any noticeable effect, but as the amount of SST is increased the number of triggered receptors increases, and so the membrane charge also goes up, as shown. There are only a certain number of available receptors on the cell's surface so when all of these are triggered regardless of how much more SST is added there will be no more increase in the membrane charge, as shown through the levelling off of the back curve at high SST concentrations. The black curve is our control, we knew this response would happen when adding SST to these cells, but it is important to check everything is as expected. The red curve is more interesting, this is the response of a special new genetically engineered peptide that is a combination of an SST molecule and a molecule of Red Fluorescent Protein (RFP). This is a protein derived from one found in some jellyfish and is a very useful as it can be genetically engineered and produced in reasonable quantities by specially adapted yeast cells. Originally, we wanted to conjugate the SST to a nano-particle, but to achieve this goal we must use some other proteins, and the danger when combining proteins is that even a slight change in their shape will destroy their activity. By producing a genetic SST-RFP molecule we can test the RFP by looking to see if there is fluorescence, and test the SST by doing the above experiment, as can be seen on the graph (red line) this new molecule also gives a similar response so we can be confident the SST is still triggering the cell, however it is a bit less effective than pure SST - hence the act of creating the new genetically engineered SST-RFP has reduced the SST quality a bit, but is still active, thus proving the whole technique to this point has worked.
This in turn leads to a small change in the charge on the cell membrane, and this is a useful tool to check if SST is entering the cell. The plot, right, shows just such an experiment. Here SST is referred to as an agonist as it is the molecule responsible for setting off the cellular responses. The bottom axis shows the number of SST molecules added to a batch of cells and the y-axis shows the recorded level of cell hyperpolarisation - the amount of charge on the membrane (this is recoded using another dye where the amount of fluorescence relates to the local charge). The curve shown (black) is typical of what is to be expected if things are working well. At low concentrations there are not enough triggered SST receptors to create any noticeable effect, but as the amount of SST is increased the number of triggered receptors increases, and so the membrane charge also goes up, as shown. There are only a certain number of available receptors on the cell's surface so when all of these are triggered regardless of how much more SST is added there will be no more increase in the membrane charge, as shown through the levelling off of the back curve at high SST concentrations. The black curve is our control, we knew this response would happen when adding SST to these cells, but it is important to check everything is as expected. The red curve is more interesting, this is the response of a special new genetically engineered peptide that is a combination of an SST molecule and a molecule of Red Fluorescent Protein (RFP). This is a protein derived from one found in some jellyfish and is a very useful as it can be genetically engineered and produced in reasonable quantities by specially adapted yeast cells. Originally, we wanted to conjugate the SST to a nano-particle, but to achieve this goal we must use some other proteins, and the danger when combining proteins is that even a slight change in their shape will destroy their activity. By producing a genetic SST-RFP molecule we can test the RFP by looking to see if there is fluorescence, and test the SST by doing the above experiment, as can be seen on the graph (red line) this new molecule also gives a similar response so we can be confident the SST is still triggering the cell, however it is a bit less effective than pure SST - hence the act of creating the new genetically engineered SST-RFP has reduced the SST quality a bit, but is still active, thus proving the whole technique to this point has worked.
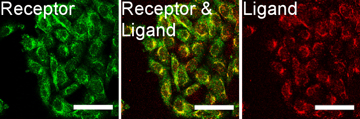 The next important test is to actually see that the new SST-RFP molecule has entered the cell, the above test showed it triggered the receptors so logically it should have, but in science it is wise never to assume anything. The images right show a test of this. Here the red colour (consider first the right most image) is our SST-RFP after incubation will some cells. As can be seen the red colours is inside the cells which is a good sign. We next add a special dye which only sticks to SST receptors (not the SST itself). This is a green colour and can be seen in the left image. When we look at both the SST and the SST receptors (middle image) we see a strong yellow colour. This is a combination of red and green and indicated that the SST and the SST receptors are in the same places, i.e., they are bound together as we had hoped. This test shows that everything is working and that, for example, the cell does not accidently destroy our new SST-RFP molecule, or see it is different and get rid of etc, and so is the final proof that if we genetically engineer an SST molecule with other functional parts, it will enter cells and work in the same way as normal SST. This is the end goal of the project, and so the final steps are to produce nano-particles covered in SST, and this research is picked up in the bio-conjugation research section.
The next important test is to actually see that the new SST-RFP molecule has entered the cell, the above test showed it triggered the receptors so logically it should have, but in science it is wise never to assume anything. The images right show a test of this. Here the red colour (consider first the right most image) is our SST-RFP after incubation will some cells. As can be seen the red colours is inside the cells which is a good sign. We next add a special dye which only sticks to SST receptors (not the SST itself). This is a green colour and can be seen in the left image. When we look at both the SST and the SST receptors (middle image) we see a strong yellow colour. This is a combination of red and green and indicated that the SST and the SST receptors are in the same places, i.e., they are bound together as we had hoped. This test shows that everything is working and that, for example, the cell does not accidently destroy our new SST-RFP molecule, or see it is different and get rid of etc, and so is the final proof that if we genetically engineer an SST molecule with other functional parts, it will enter cells and work in the same way as normal SST. This is the end goal of the project, and so the final steps are to produce nano-particles covered in SST, and this research is picked up in the bio-conjugation research section.
Papers relating to this work:
 Non-specific cellular uptake of surface-functionalized quantum dots
Non-specific cellular uptake of surface-functionalized quantum dots
T.A. Kelf, V.K.A. Sreenivasan, J. Sun, E.J. Kim, E.M. Goldys and A.V. Zvyagin
Nanotechnology.21, 285105 (2010)
 Pharmacological Characterization of a Recombinant, Fluorescent Somatostatin Receptor Agonist
Pharmacological Characterization of a Recombinant, Fluorescent Somatostatin Receptor Agonist
V.K.A. Sreenivasan, O.A. Stremovskiy, T.A. Kelf, M. Heblinski, A.K. Goodchild, M. Connor, S.M. Deyev, and A.V. Zvyagin
Bioconjugate. Chem. 22, 1768-1775 (2011)
 Barstar:barnase - a versatile platform for colloidal diamond bioconjugation
Barstar:barnase - a versatile platform for colloidal diamond bioconjugation
V.K.A. Sreenivasan, E.A. Ivukina, W. Deng, T.A. Kelf, T.A. Zdobnova, S.V. Lukash, B.V. Veryugin, O.A. Stremovskiy, A.V. Zvyagin, and S.M. Deyev
J. Mat. Chem.21, 65 (2011)
 A modular design of low-background bioassays based on a high-affinity molecular pair barstar:barnase
A modular design of low-background bioassays based on a high-affinity molecular pair barstar:barnase
V.K.A. Sreenivasan, T.A. Kelf, E.A. Grebenik, O.A. Stremovskiy, J.M. Say, J.R. Rabeau, A.V. Zvyagin, S.M. Deyev.
Proteomics. 13, 1737-1443 (2013)